Last revised:
Wednesday, March 8, 2000
Ch. 11 (p. 212-225) in Prescott et al, Microbiology, 4th Ed.
Note:
These notes are provided as a guide to topics the instructor hopes to cover during lecture. Actual coverage will
always
differ somewhat from what is printed here. These notes are not a substitute for the actual lecture!
Copyright 2000. Thomas M. Terry
- Every living organism has DNA = cell database. Bacteria have single chromosome (circular in all except Borrelia burgdorferi, cause of Lyme disease), no nucleus.
- View TEM of bacterium, illustrating DNA (marked n) in cell cytoplasm. Cell is a dividing Neisseria gonorrhoeae, cause of gonorrhea.
- View electron micrograph of isolated bacterial DNA
- Central dogma: information is encoded in DNA. To express this information, RNA is transcribed with same coding, then translated into amino acid sequence which folds to form active proteins.
- View animation of central dogma (Note: shockwave plug-in required)
- DNA encodes two types of molecules:
- database for protein structure (access by sequential transcription & translation)
- database for needed t-RNA, r-RNA molecules (access by transcription alone)
- Several bacterial genomes have been completely sequenced. Some chromosome sizes are listed in the table, along with sizes of yeast and human DNA for comparison.
| Organism | Domain | Chromosome size (base pairs) | Predicted polypeptide coding regions |
| Mycoplasma genitalium | Bacteria |
0.58 Million bp |
470 proteins |
| Hemophilus influenzae | Bacteria |
1.83 Million bp |
1740 proteins |
| Methanococcus jannaschii | Archaea |
1.66 Million bp |
1682 proteins |
| Escherichia coli | Bacteria |
4.64 Million bp |
4288 proteins |
| Largest yeast chromosome now mapped | Eukarya |
1.55 Million bp |
? |
| Entire yeast genome | Eukarya |
15 Million bp |
? |
| Smallest human chromosome (Y) | Eukarya |
50 Million bp |
? |
| Largest human chromosome (1) | Eukarya |
250 Million bp |
? |
| Entire human genome | Eukarya |
3 Billion |
? |
- View TIGR database of published and in progress gene sequences
- E. coli cell is often used as "model organism". Cell ~ 2 micrometers in length; has single circular DNA
chromosome. Can be measured in various ways:
- ~1400 micrometers in length
- 4700 kilobase pairs
- 2 x 109 daltons in mass
- DNA prompts many intriguing questions. For example
- how does cell manage a
giant ball of string, which must be identically copied prior to replication,
without generating hopeless tangle?
- Many genes are not expressed most of
the time, only under certain circumstances. How does cell regulate this
expression?
- Any damage to DNA is likely to be lethal, since there is no
"backup" copy of the database. How does cell detect and repair damage?
- Unit of atomic weight is the dalton. A Hydrogen atom weighs 1 dalton; an Oxygen atom weights 16 daltons.
- Often refer to DNA by weight: e.g. virus DNA weighs in the range of millions of
daltons, bacterial DNA weighs several billion daltons.
- Average nucleotide weight is ~ 330 daltons; one base pair has average mol. wt.
of 660 daltons. Divide weight of double-stranded DNA by 660 to get approx. # of
base pairs.
- Example: E. coli weighs 2.5 billion daltons. Divide by 660 = roughly 4
million base pairs
- Since most DNAs have anywhere from thousands to billions of base pairs, use
units of kilobase pairs (Kbp) = 1000 bp. So E. coli in above example would have
roughly 4000 Kbp (pronounced kilo base pairs).
- Another common measure is Million base pairs (Mbp). E. coli has 4 Mbp of DNA.
- Note: single-stranded RNA or DNA would be measured in Kb (kilo bases), not base
pairs.
- DNA is tightly coiled = supercoiled. Coiling is maintained by family of
enzymes known as topoisomerases. Eg., DNA gyrase (topoisomerase II)
induces supercoiling; topoisomerase I causes relaxation of supercoil. ATP
required for supercoiling.
- View electron micrograph showing supercoiled DNA (from Institute for Molecular Virology. University of Wisconsin - Madison)
- First enzyme isolated by Kornberg (----> Nobel prize): DNA polymerase I.
- First Reaction:
[dATP, dCTP, dGTP, dTTP] -------(DNA polymerase,
Mg++, template DNA)--------> new DNA + P~P (pyrophosphate)
- Second reaction:
P~P ------(pyrophosphatase)-----> 2 Pi (inorganic
phosphate)
- Note 1: DNA is the one molecule the cell can absolutely not afford to see
broken down!!!!
Reaction 2 is necessary to keep conc. of P~P vanishingly
small; otherwise mass action law would promote slow breakdown of DNA towards
equilibrium state.
- Note 2: energy for forming new sugar-phosphate bond comes from splitting a
high-energy phosphate bond as P~P is removed. This always occurs at free 3'-OH
group on deoxyribose (and on ribose in RNA synthesis). All nucleic acids grown
by addition at 3'-end, not at 5'-end. Often referred to as 5' -----> 3'
synthesis. (See handout).
- View DNA replication (from The Biology Place)
- After Kornberg's discovery, questions arose as to whether polymerase I could in
fact be solely responsible for replication? Evidence suggested greater role in
repair. Two more enzymes later isolated: DNA polymerase II and DNA polymerase III. Turns out DNA
polymerase III is the principal replication enzyme, though polymerase I has a
role also.
- Other enzymes and proteins involved:
- DNA helicase. This unwinds DNA just in front of opening replication
fork (otherwise DNA would quickly tangle). Uses ATP, makes single-stranded cut,
allows one strand to swivel freely around the other.
- Single-stranded DNA binding proteins. These bind to separated DNA
strands, prevent from base-pairing back together
- RNA primase. DNA polymerase III cannot start a growing chain from
scratch; needs a short primer (a few nucleotides) to add to. This is carried
out by DNA-dependent RNA primase, makes very short piece of RNA by base-pairing
RNA nucleotides with template DNA.
- DNA polymerase III. This adds new nucleotides at free 3'ends of
growing chain, uses base-pairing rules to insert complementary nucleotides (A
opposite T, G opposite C, etc.) Can keep on adding indefinitely for millions
of nucleotides if not blockage.
- DNA polymerase I. This removes RNA primers, fills in gaps by base
pairing, inserts new DNA nucleotides to replace RNA primer. Basically a repair
enzyme, but required here.
- DNA ligase. A "sealing" enzyme, required to join any gaps where
adjacent nucleotides on one strand have not been covalently joined. In
bacteria, use phosphate bond of NAD+ as energy source; some ligases
use ATP for energy.
- Note: many gaps result on lagging strand (see below), so lots of need for
enzymes (5) and (6).
- Since two strands in DNA are antiparallel, new DNA must be synthesized in
opposite directions on the two template strands.
- But overall, DNA must unwind in one direction (at replication fork), overall
DNA synthesis has one direction.
- No problem for the strand growing in same direction as unwinding = leading
strand. Can make one long, continuous piece of DNA
- Big problem for strand growing in opposite direction to unwinding = lagging
strand; must grow away from unwinding. As new template is opened up by DNA
unwinding, will have to start a new copy.
- In fact, just this situation was discovered experimentally by Okazaki; found
many short DNA fragments newly synthesized from lagging strand = Okazaki
fragments. Must be joined together by DNA ligase to make continuous DNA
strand.
- View animation of bidirectional strand replication (from The Biology Place)
- Curious fact: E. coli can grow with generation time as low as 20 minutes
at 37o C, yet complete replication of chromosome takes much longer (even at
750-1000 base pairs per second, there's an awful lot of DNA!). How?
- Answer 1: DNA chromosome replicates bidirectionally (two replicating forks each
proceeding in opposite directions). So can replicate the whole chromosome in
half the time if only unidirectional synthesis. Generates "theta structures"
which resemble Greek letter theta during replication.
But still would take
~40 minutes! How to grow cell in 20 minutes?
- Answer 2: Second round of DNA replication is initiated long before first round
is finished. In exponential growth, cells can accumulate 1 or two rounds of DNA
before division occurs. Possible to find up to 4 identical chromosomes in a
single cell! (But not for long, cell is primed for division, eventually as
nutrients diminish will wind up with only 1 chromosome per cell).
- View bidirectional replication in DNA, with multiple initiation sites. (Phage lambda, from the Institute for Molecular Virology. University of Wisconsin - Madison)
- Any damage to DNA would be lethal. Cells can spend more energy repairing DNA
than synthesizing it. Will discuss later.
- Also proofreading is necessary. When new DNA is synthesized, occasional errors
in base pairing occur. If not corrected, could lead to mutations, loss of
functions, loss of competitiveness, evolutionary weeding out.
Proofreading carried out by DNA polymerases III and I; if base mismatch
spotted, cut out new bases (keep track of which is template strand and which is
new strand during replication), resynthesize copy strand from that neighborhood
of template.
- Circular DNA elements, always double-stranded DNA, Supercoiled
-
Can occur in as few as 1 copy per cell (single copy plasmids) to as many as
several dozen (multicopy plasmids).
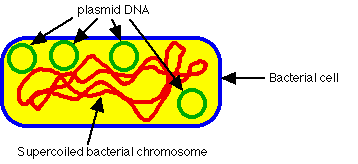
-
Variable sizes; small plasmids about 0.1% size of host chromosome, large
plasmids can be as much as 10% the size of host chromosome. Smaller plasmids
have few genes (30 or less). Size ranges from 1000 bp (1 kbp) to 1000 kbp.
-
Ubiquitous; almost all cells isolated in nature carry plasmids, often more than
one kind. (In E. coli alone, more than 300 different plasmids isolated.)
- View EM of plasmid DNA
-
Have a replicon (origin for DNA replication), number of copies per cell
regulated. Large plasmids typically only 1-5 copies/cell (stringent control);
small plasmids ~10-50 copies/cell (relaxed control)
-
Many plasmids are incompatible; if one is present, cell cannot support another
plasmid of same compatibility group.
-
Not essential to cell under all circumstances; can be "cured" by agents that
impair DNA replication ----> cured cell lacking plasmid. Can be
spontaneously lost over time unless some selection makes plasmid valuable to
cell.
-
Extend range of environments in which a cell can live (e.g., by degrading
antibiotics, or providing enzymes for digestion of novel catabolites).
- Antibiotic resistance genes (enzymes that modify or degrade antibiotics) --
plasmids with these genes are called R factors
-
Heavy metal resistance (enzymes that detoxify metals by redox reactions)
-
Growth on unusual substrates (enzymes for hydrocarbon degradation, etc.)
-
Restriction/modification enzymes (protect DNA, degrade unprotected DNA)
-
Bacteriocins (proteins toxic to other bacteria lacking the same plasmid)
-
Toxins (proteins toxic to other organisms; e.g. humans) -- called virulence
plasmids. Some Examples:
- Staph aureus virulence factors: coagulase, hemolysin, enterotoxin, others
- pathogenic E. coli strains: hemolysin, enterotoxin
-
Proteins that mediate plasmid transfer to uninfected strains
Structure:
- Components:
- Ribose
- Phosphate
- Purine bases A, G; Pyrimidine bases C, U
- View comparison of RNA and DNA (from The Biology Place)
- U has same base pairing properties as T (forms U=A base pairs)
- RNA is not double stranded (except in some viruses)
- RNA can have extensive "hairpin" loops
- RNA can have modified bases (after transcription) -- find unusual bases such as inosine, pseudouridine. Still have base pairing properties. Probably contribute to stability of molecule, aren't recognized by RNases which break down unmodified RNA. Messenger RNA has half-life of only 3 minutes in E. coli, so cell must constantly make new messages to make new proteins-- allows rapid adaptation to new environments.
Types of RNA
- messenger RNA -- carries codons to RNA
- ribosomal RNA -- part of ribosome structure, catalyzes peptide bond formation
- transfer RNA -- small RNAs (about 60 diff. types in E. coli), transport amino acids to ribosome for incorporation into growing polypeptides
Transcription
- View animation of transcription (requires password to The Biology Place)
- Requires enzymeRNA polymerase
- opens up DNA helix for short stretch (~ 15 base pairs)
- View RNA polymerase attached to short DNA molecule (electron micrograph)
- selects one of two strands as template strand
- RNA synthesized in 5' to 3' direction
- Promoters: sites on DNA that are recognized as "start" signals for RNA synthesis. Typical promoter has "TATA..." base sequence (consensus sequence -- some variation) at -10 bases (upstream from actual site of RNA synthesis).
- role of sigma factor-- binds to RNA polymerase core enzyme, recognizes promoter. Dissociates
- View model of TATA-binding protein
- Note: Bacteria can have more than one sigma factor, recognize different types of promoters.
- For example: in E. coli, when cells enter stationary phase, new sigma factor made. This allows RNA polymerase to recognize some new genes with different promoter, make RNA and then proteins which can stabilize cell during starvation, allow it to maintain quasi-dormant state until new nutrients become available.
- Another example: spore-forming cells activate different sigma factor to recognize new sets of genes involved in spore formation.
- Terminators: can be stem-loop structures with poly-U runs, or certain sequences recognized by rho factor.
Antibiotic effects
- rifamycin (prokaryotic): affects beta subunit of RNA polymerase
- actinomycin: (acts on both prokaryotic and eukaryotic cells): binds to DNA at GC pairs, blocks attachment of RNA polymerase
Take a Self-Quiz on this material
Return to Lecture Index
Return to MCB 229 Course Resources page

