|
|
Quiz Me! |
Microbial genetics: general principles |
|
|
|
Lecture Index | ||
|
|
Course Resources page |
Last revised: Wednesday, March 29, 2000
Ch. 13 in Prescott et al, Microbiology, 4th Ed.Note: These notes are provided as a guide to topics the instructor hopes to cover during lecture. Actual coverage will always differ somewhat from what is printed here. These notes are not a substitute for the actual lecture!Copyright 2000. Thomas M. Terry
Unique features of bacterial genetics
- Single genome per cell
- Fast growth rates ---> rapid results
- Enormous numbers of offspring; even the most improbable events can occur at significant rates
- Genome can be sequenced in reasonable time
Terminology
Mutation and mutation rate
- mutation = heritable change in DNA (or RNA for RNA viruses)
- can come about as a result of single base change, multiple base changes, even addition or deletion of large amounts of DNA
- effects of a mutation range from being
- undetectable (e.g., base change leads to a same amino acid being specified, or slightly different amino acid with no effect on protein structure or activity)
- slight (e.g., amino acid substitution leads to altered protein, slight change in active site)
- severe (e.g., amino acid substitution alters active site, enzyme no longer binds substrate)
- lethal (e.g. mutation affects a critical enzyme such as RNA polymerase)
Wild-type and mutant
- wild-type is capable of full range of metabolic activities found in type specimens. Ex: wt E. coli can manufacture all 20 amino acids from single C-source, manufacture all vitamins, fatty acids, vitamins, etc.
- Mutant could be defective in synthesis of some substance, e.g. amino acid leucine (leu- strain); would have to be fed leucine in order to grow.
Phenotype and genotype
- Phenotype = what is observed. Ex: red pigmented colony, resistant to antibiotic, requirement of leucine for growth
- Genotype = symbolic representation of gene responsible for phenotype. Ex: leu-, strR
- Typically use 3-letter codes for genotype.
Naming bacterial strains
- Every isolate should be identified by strain designation; e.g. E. coli K12, E. coli B
- Don't name wt genes, only mutants: e.g. E. coli B leu- thr- lac- penR (but not E. coli K12 leu+ thr+ lac+ penS)
- Collections of strains are kept.
- Major U.S. source is ATTC (American Type Culture Collection); can order any strain for minimal fee.
- ATTC maintains the following frozen or freeze-dried stocks: (number of species in parentheses)
- Algae (120)
- Bacteria (14400)
- Fungi (20200)
- Yeasts (4300)
- Protozoa (1090)
- Cell lines: animal (2300)
- Cell lines: plant (25)
- Viruses: animal (1350)
- Viruses: plant (590)
- Viruses: bacteria (400)
- Yale Univ. keeps the definitive E. coli collection (in freeze-dried form in envelopes); can search computer database, identify specific strains to order.
Mechanisms of Mutation
Nucleotide substitution (base change)
- "spontaneous error": can arise from diverse sources; don't know exact cause of any specific mutation. Errors in base pairing occur, even after proofreading, with frequency of 1 in 107 to 1 in 108.
- base analogs: compounds like 5-bromouracil look like Thymine, get incorporated into DNA during replication. However, when serving as templates, they don't always form the "correct" match of A, instead sometimes pair with C.
- nitrous acid: causes deamination; resulting bases can cause incorrect base pair matches
- UV radiation: causes fusion of adjacent thymine residues in same strand ---> thymine dimers. Can be repaired, but if not will cause inconsistent base insertions during replication.
- Note 1: all of these approaches often produce multiple mutations in different genes
- Note 2: these mutations can potentially be reversed by a second mutation which substitutes the original base for one altered to produce initial mutant. Such mutants are called revertants. Most mutations can revert with some frequency, even if slight.
Frameshifts (by insertion or deletion of a base pair)
- Certain chemicals called intercalating agents can slip into DNA double helix between base pairs, induce mutations that result in extra bases being added.
- Resulting genetic code now has extra base, will be frame shifted at some point. Protein is made, but typically garbage, has no relation to original protein, often has "stop" codon earlier than wt gene, causes truncated garbage proteins.
- Acridine dyes are good intercalating agents, cause frameshift mutations.
- Note: to get frameshift revertants, typically have to use a frameshift mutagen.
Deletion mutations
- Can result from ionizing radiation, other treatments that cause double-stranded breaks. During DNA repair, section of DNA can fail to get reintegrated ----> deletion of dozens, maybe thousands of base pairs of DNA.
- Deletion mutants cannot revert. Very stable mutants.
Insertion mutagenesis
- Transposons are movable genetic elements, flanked by insertion sequences.
- When transposon moves, can insert itself within a structural gene.
- Example: Like taking sequence XYYWWLALL and moving in into coherent phrase; FOURSCORE AND SEVEN
.... Result: FOURSCXYYWWLALLORE AND SEVEN .... this is gibberish, destroys sense of a word. Similarly, inserted DNA will be transcribed, and disrupt the normal protein.- Value of transposon mutagenesis: can get a single insertion (rather than a cluster of mutants as in chemical mutagenesis); can actually find site of mutation on gels after digesting DNA with restriction enzymes.
Types of Mutants and selection strategies
Colony morphology
- Easy to detect (e.g. colony smooth rather than slimy); often reflect changes in genes affecting cell surface
Resistant mutants
- Easy to select; add an antibiotic or a virus, look for zones where most cells are inhibited, a few mutants can resist agent and will grow.
Auxotrophs
- Auxotroph = mutant that cannot grow on minimal medium, requires certain supplement(s).
- Many auxotrophs have been isolated. Very useful in figuring out metabolic pathways
- Example: ser- auxotroph, can't grow without added serine. How to select such a mutant?
Naive selection procedure for auxotrophs
- Screen for mutants: spread cells on a plate containing serine so mutants and wt will both grow (can't yet tell which is which).
- Locate isolated colonies. Pick each colony, transfer to two plates:
- no added serine
- + added serine
- Locate colonies so can compare same colony on each plate.
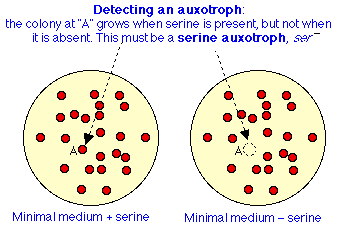
- Now incubate. If colony grows on plate (2) but not (1), it is a desired mutant. Can't pick from (1) (it's not there, remember?) but can pick from comparable site on plate 2.
- Store this colony, give it a mutant number, repeat as long as needed to get reasonable number of mutants.
- Problem: mutant frequency is low. Might have to screen 10,000 plates just to find a single mutant. Too much work!! Need better odds.
- Enrichment culture. Need to find a way (enrichment culture) to enrich for serine auxotrophs in original culture. But how? serine auxotrophs can do everything wild type can do except grow in serine.
- Solution (devised by Joshua Lederberg): penicillin selection. Penalize cells that can grow, reward cells that can't grow.
Penicillin selection procedure for auxotrophs
- mutagenize culture to increase mutation frequency
- grow cells in minimal medium supplemented with 19 amino acids (but not serine), vitamins, other biosynthetic needs for which auxotrophs other than serine have needs. Result: all cells, both wt and other auxotrophs, will grow. But serine auxotrophs can't grow.
- when cells are happily in exponential growth, throw in penicillin (or other cell wall antibiotic). Will lead to wall damage in all growing cells, but not in serine auxotrophs.
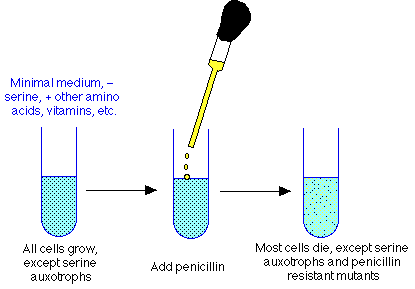
- after lysing cells, plate survivors on plates containing serine, use two plate screening technique described above. Should enrich for serine auxotrophs considerably.
- Note: same type of procedure can be used to select any auxotroph. In practice, try to find a lot of independently isolated mutants, then study and compare them (some will be in same gene).
Conditional lethal mutants
- For any gene, possible to get mutations that affect protein folding. Some of these will cause protein to denature at modestly high temperatures (e.g. 42o C), whereas protein will be stable at cooler temps (30o C). These are called temperature sensitive mutants, one example of a conditional lethal mutant (lethal under one set of conditions, not under another)
- If such mutations occur in gene absolutely required for cell survival, then at higher (restrictive) temperature, protein will unfold and cell will die. At lower (permissive) temperature, protein folds normally and cell can grow
- Easy to select:
- mutagenize
- grow cells at 30o, plate out colonies.
- test each colony for growth at 30o and at 42o.
- Pick colonies that survive 30o but die at 42o.
- Result: can isolate a large class of mutants that are temperature-sensitive. These mutants are entirely distinct, except that all affect proteins critical for survival.
- Study individual ts mutants. Can discover many genes and their protein products involved in critical cell processes such as cell division, DNA replication and separation, RNA synthesis, etc.
Ames test
An application of power of bacterial genetics to help screen for substances that might cause cancer.Carcinogens
- Many substances can cause cancer: large number of chemicals, radiation, etc.
- Wide variety, but common denominator in general is that almost all carcinogens cause mutations in DNA. When critical cell targets regulating cell division are mutated, result is cancer. Can occur in any tissue.
Difficulties with carcinogen testing
- NIH has protocols for testing suspected carcinogens. Requires special strains of inbred animals (genetically homogenous), different dose levels, multiple repeats, statistical analysis, various techniques for assessing presence or absence of tumors in each animal.
- Very expensive: can cost anywhere from hundreds of thousands to millions of $$, take from 6 months to 2 years to test
- Thousands of new chemicals are introduced to U.S. industry each year, find their way into cosmetics, foods, drugs, consumer products. Impossible to screen most of these for possible carcinogen activity.
- But could screen for mutagenic activity; take chemicals that show up positive, screen those for carcinogenic potential. A very efficient strategy; bacteria are cheap, quick.
Design of the Ames test
- Bruce Ames developed test using histidine auxotrophic mutants of Salmonella (cousin of E. coli)
- Ames tester strains are his- point mutants (possibility of reversion mutation is there).
- Test design:
- Control Plate: spread ~ 108 his auxotrophs on a plate containing minimal medium, lacking histidine. Result: cells won't grow, except for occasional revertant spontaneous mutant (at 1 in 107 rate, expect ~ 10 mutants/plate).
- Experimental plate: spread same cells on similar plate, add a filter disk soaked in test chemical solution. If chemical is mutagenic, will diffuse into agar, will see increased number of mutants surrounding the disk.
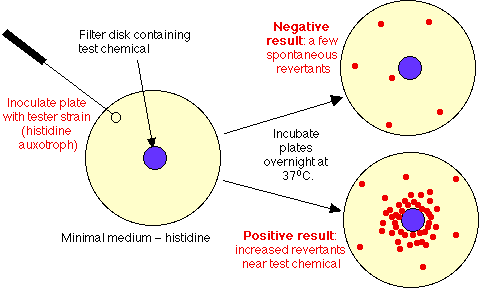
- Note: actually this test as just described will miss many chemicals that are mutagens in animals. Why? In animals, chemicals are detoxified in liver, often by many chemical steps. In process, some chemicals which are not initially mutagenic are converted into mutagens. To expand scope of Ames test, must add preparation of liver enzymes (made by grinding up fresh animal liver, centrifuging out debris) = liver microsomal fraction. With this addition, many more chemicals show up as Ames positive.
- Economics of Ames test: costs only a few $100, instead of millions. Takes only a couple of days, instead of a year or more. ~90% of chemicals that test positive for mutagenesis have been found to be carcinogenic in animals. There are some carcinogens that don't show up as mutagens on Ames test, so it's not foolproof, but a good screen.
- Many industries now routinely use Ames test as screening for new products, will not develop products further if positive test (good practice in the age of soaring liability costs).
Take a Self-Quiz on this material
Return to Lecture Index
Return to MCB 229 Course Resources page