|
|
Quiz Me! |
Regulation of Metabolism |
|
|
|
Lecture Index | ||
|
|
Course Resources page |
Last revised: Thursday, March 16, 2000
Ch. 12 (p. 237-250) in Prescott et al, Microbiology, 4th Ed.Note: These notes are provided as a guide to topics the instructor hopes to cover during lecture. Actual coverage will always differ somewhat from what is printed here. These notes are not a substitute for the actual lecture!Copyright 2000. Thomas M. Terry
Metabolic Channeling
- If groups of enzymes and metabolites are separated from others, metabolism will be more efficient.
- This occurs commonly in eukaryotes with multiple compartments; less so in prokaryotes
Regulation of enzyme activity
- Most enzymes are fully active; activity depends on conc. of substrate
- But some enzymes can be turned on and off. These are called allosteric enzymes, with active site for substrate and effector site for regulator
- Example: pathway for synthesis of amino acids proline and arginine. Both are made from glutamic acid, after several enzyme-catalyzed reactions. First enzyme in each pathway is inhibited by end-product. So if pro accumulates, shuts off synthesis of pre-existing proline; arg can still be made.
- Note: this is different from gene regulation; pro can suppress transcription (effect won't show up for several minutes), also turn off existing enzyme pathway (effect shows up immediately).
Regulation of gene expression in prokaryotes
Many genes occur in operons
- several structural genes (for protein) controlled by a single promoter
- mRNA is polycistronic: has multiple start & stop signals, codes for multiple polypeptides
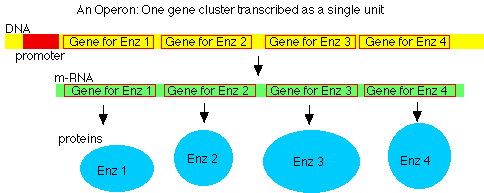
- In some cases, transcription depends on specific DNA region, the operator, which is near or overlapping the promoter.
- Operator site can bind repressor proteins, controlled by some effector molecule
Transcription & Translation are coupled
- Prot. synthesis begins before transcription ends
- multiple ribosomes bind to each mRNA
- All control at level of transcription
- once m-RNA is made, it is translated.
Some genes are expressed constitutively
- Always translated when cells synthesizing protein
- Examples
- ribosomal genes
- genes for replication & transcription
- genes for central metabolism enzymes
- Note: some proteins made from constitutive genes occur at much higher levels than others. Why? Strong vs. weak promoters.
Some genes are inducible: transcribed only in presence of inducer
- Classic example: the lac operon
- gene is regulated by negative control; in absence of specific repressor, gene is transcribed just like constitutive gene. In order to regulate, must add specific block. Must say "no"; otherwise gene is not down-regulated.
- View Animation of negative control from Cornell University
- Lactose = milk sugar, disaccharide made of galactose + glucose. In order to metabolize lactose, cells must produce enzyme ß-galactosidase, split lactose into galactose + glucose
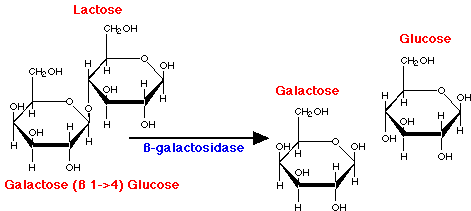
- Observation: add lactose to cells: within minutes, ß-galactosidase enzyme appears, also lac permease in membrane, and a third protein, transacetylase. Level of ß-galactosidase enzyme can accumulate to level of 10% of cytoplasmic protein!
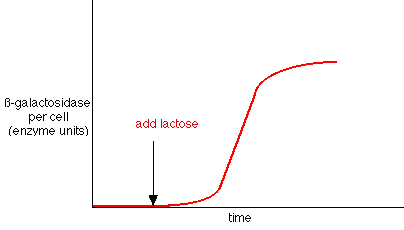
- Explanation: in absence of lactose (= inducer), lac repressor blocked operator site.
- Lac repressor is allosteric protein. Coded for by another region of DNA (constitutive gene, weak promoter, low level of expression)
- View animation of lac repressor (access to The Biology Place required)
- Effector molecule = Inducer is lactose (actually allolactose, or analog such as IPTG) binds to repressor protein, repressor released, RNA is made, all genes turned on as unit
- View animation of lactose induction (access to The Biology Place required)
- In lab, MaConkey agar used to detect lac mutants. If lactose is used (lac genes on), fermentation occurs in colony, acid produced, color indicator turns pink. Pink colony = lactose utilized. But if lactose genes don't work (mutant), colonies grow but stay white.
- To review these concepts, look at Cornell University's negative control explanation.
Some genes are repressible; transcription is shut down in presence of repressor
- Very similar to inducible genes, except for type of gene affected and type of effector molecule
- Inducible genes turned on when some catabolite enters environment (e.g. a sugar). Repressible genes produce normal cell molecules (e.g. amino acids), but can be turned off when excess product accumulates; e.g. when environment supplies amino acid. Sensible: why spend E making it if it's free?
- Ex: arginine synthesis enzymes: external arg represses further enzyme synthesis for arg operon (but other enzymes continue being synthesized). Repressor + arg binds to DNA at arg operator, blocks RNA polymerase.
- Also an example of negative control.
Levels of regulation
- Negative control
- Discussed above: both inducible and repressible genes are examples of negative control
- Positive control
- So far, assumed RNA polymerase always able to transcribe unless blocked by repressor protein (negative control).
- But many genes are not transcribed unless some protein binds to DNA, facilitates RNA Polymerase activity. This is Positive control (nothing happens unless you say "Yes".)
- View animation of positive control from Cornell University. (Requires shockwave plug-in).
- Global Regulation
- Catabolite Repression.
- Synthesis of variety of different enzymes involved in catabolism are inhibited if glucose is present. (Can also be called "glucose effect"). Flagellar synthesis also controlled by catabolite repression.
- View animation of catabolite repression from Cornell University (requires schockwave plug-in)
- Diauxic growth curve illustrates the effect
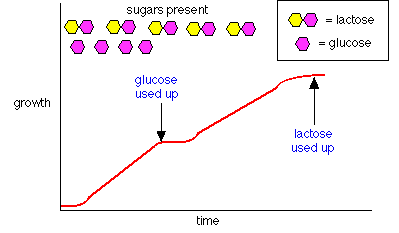
- How does it work? combination of positive & negative control.
- Already saw negative control system: need inducer to get rid of repressor
- BUT also need activator protein (called Catabolite Activator Protein, or CAP)
- CAP is allosteric protein, only binds to DNA if cAMP is present. cAMP is made from ATP by enzyme adenyl cyclase, but is inhibited by glucose.
- Cell signal: glucose high, no cAMP. CAP doesn't bind, not positive control for dozens of operons. But if glucose conc. falls low, cAMP produced, CAP binds, now many operons can be used (depending on other inducers & repressors).
- View CAP protein bound to DNA, from Cornell University.
Group Practice
- Alternative Sigma factors
- In E. coli most promoters recognized by sigma70. But if cells experience severe temp. shock (e.g. heat), new set of genes turned on to "rescue" cells from disaster.
- New sigma factor is made, recognizes different set of genes with different promoters.
Gene organization in Prokaryotes vs. Eukaryotes
Regulation in prokaryotes and eukaryotes
PROKARYOTES EUKARYOTES DNA geometry single circular molecule multiple linear molecules Replication origin (ori) one per DNA molecule many per chromosome DNA replication rate 750-1000 bp/sec 50-100 bp/sec. Rate is 10x times slower than bacteria. However, since eucaryotic chromosomes have many ori sites, overall time required for DNA replication can actually be less than for bacteria (which are limited to one ori site). Cell's use of DNA almost all DNA codes for protein large regions of noncoding DNA Gene organization many operons (one promoter + several structural genes)
genes lack introns; DNA is colinear with polypeptide (rare exceptions) no operons (one promoter + one structural gene)
genes contain introns; must be removed in nucleus before mRNA is formed
some large polypeptides cleaved into multiple active proteins RNA polymerase 4 subunits in single "core enzyme", plus sigma or rho factors
Archaea have 8-10 subunits, single enzyme 10-12 subunits, 3 different enzymes mRNA organization many mRNAs have multiple "start" and "stop" signals; code for multiple proteins (polycistronic) mRNAs have one "start" and "stop" signal; code for one protein (monocistronic) Control of RNA synthesis some genes constitutive some genes inducible some genes repressible some genes cryptic some genes constitutive
inducible genes regulated by complex set of activator proteins Fate of RNA translated immediately (coupled transcription-translation short half-life not translated immediately 5' methyl Guanosine "cap" and 3' poly-A "tail"capped" are added to new RNA processed in nucleus to remove introns splicing is catalyzed by RNA (ribozyme) rather than protein enzyme exported from nucleus to cytoplasm long half-life Protein synthetic rate 350-400 amino acids/min
70S ribosomes; 30S + 50S subunits 50 amino acids/min
80S ribosomes; 40S + 60S subunits "Start" amino acid (AUG codon) N-formyl-methionine (but methionine in Archaea) methionine antibiotic effects
- rifamycin (prokaryotic); affects beta subunit of RNA polymerase
- amanitin (eukaryotes); affects only mRNA polymerase of eukaryotes
Take a Self-Quiz on this material
Return to Lecture Index
Return to MCB 229 Course Resources page