|
|
Quiz Me! |
Microbial Taxonomy & Evolution |
|
|
|
Lecture Index | ||
|
|
Course Resources page |
Last revised: Tuesday, March 28, 2000
Ch. 19 in Prescott et al, Microbiology, 4th Ed.Note: These notes are provided as a guide to topics the instructor hopes to cover during lecture. Actual coverage will always differ somewhat from what is printed here. These notes are not a substitute for the actual lecture!Copyright 2000. Thomas M. Terry
Introduction: Two different possible goals
(1) Convenient ordering scheme. Use of a "key"
- Phenetic system: groups organisms based on mutual similarity of phenotypic characteristics. May or may not correctly match evolutionary grouping.
- Example: Group flagellated (motile) organisms in one group, non-motile organisms in another group.
(2) Scheme displaying evolutionary relationships.
- Phylogenetic system: groups organisms based on shared evolutionary heritage.
- Example: Mycoplasma (no wall) and Bacillus (walled Gram+ rods) are not obviously similar, would not be grouped together phenetically. But evolutionarily they are similar, more so than either to Gram- organisms.
Numerical Taxonomy
- Use a variety of characteristics: e.g., Gram stain, cell shape, motility, size, aerobic/anaerobic capacity, nutritional capabilities, cell wall chemistry, immunological characteristics, etc.
- Relies on similarity coefficients
- If use 10 characteristics, then match organisms.
- Ex. A and B share 8 characters out of 10: similarity coefficient Sab is 8/10 = 0.8
- Can use many such values to establish similarity matrix
- Dendrograms help display this information clearly.
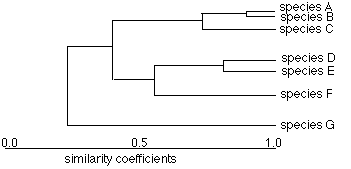
Note: dendrogram is just a graphical display of similarity coefficients; but one often assumes that these are representative of a deeper evolutionary relationship. This may or may not be legitimate conclusion, depending on the traits used. The diagram below is a hypothetical evolutionary diagram, superficially similar to a dendrogram but actually quite different, since it seeks to portray an accurate picture of how and when organisms diverged from common ancestors over time.
To get accurate phylogeny, must decide which characteristics give best insight. DNA and RNA sequencing techniques are considered to give the most meaningful phylogenies.
Terminology
strain:
- descended from a single organism
- different isolates may be same species but are different strains; often have slight differences
type strain:
- the first strain isolated or best characterized
- kept in collections; e.g., ATCC (American Type Culture Collection) maintains the following frozen or freeze-dried stocks: (number of species in parentheses)
- Algae (120)
- Bacteria (14400)
- Fungi (20200)
- Yeasts (4300)
- Protozoa (1090)
- Cell lines: animal (2300)
- Cell lines: plant (25)
- Viruses: animal (1350)
- Viruses: plant (590)
- Viruses: bacteria (400)
many similar strains = species
strain, species, genus, family, order, class, division, kingdomExample:
Genus: Escherichia
Species: coli
Family: Enterobacteriaceae
Class: Scotobacteria
Division: Gracilicutes
Kingdom: Procaryotae
"Classical Taxonomy"
Bergey's Manual: a phenetic system
- Uses both morphological and Physiological characteristics
- Very practical system. Use successive "key" features to narrow down identification
- Ex. Gram + or -? Then shape? Then motile or not? etc. Eventually only a few organisms match the process of elimination.
"Molecular Taxonomy"
Basic assumptions
- Genes mutate randomly
- Many mutations are "neutral" -- do not lead to any obvious disadvantage to the strain.
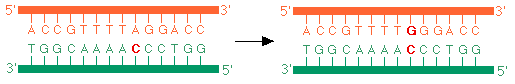
- Once a mutation is established, all progeny of parent cell carry that particular mutation. For example, in figure below, if template "A" is erroneously replicated to a "C" in the opposite strand instead of T, then one generation later the error will be "locked into place", and all progeny with that DNA will be forever altered (unless a reversion mutation occurs at some later time).
- Two organisms that differ by only a few bases have diverged more recently in evolutionary time than organisms that differ by more bases.
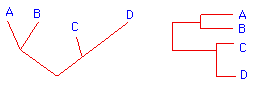
- The following example shows how an evolutionary tree is constructed for four hypothetical organisms whose DNA sequence in one homologous region is known.
AACGTCGAAA (Organism A) AACCTCGAAA (Organism B) AGGCTAGAAA (Organism C) AGGCTAGTAA (Organism D)Organism A and B differ by one base substitution. C and D also differ by one base substitution. But A and C differ by three substitutions, and A and D by four. B and C differ by three substitutions, and B and D also by four. In terms of evolutionary history, A and B appear to be very similar,as do C and D. A-B and C-D are more distantly related.Computers excel at taking such data and creating trees that accurately illustrate the divergence between different organisms, with linear distance being proportional to the number of accumulated errors. Here are a couple of ways a computer could represent the separation between these four organisms:
Molecular "clocks"
- Attempt to use basic assumptions to establish history of evolutionary lineages
- Over long periods of time, assume mutations occur with roughly predictable frequency
Use of 16S RNA sequence homology
- 16S RNA is found in small ribosomal subunit (30S) of procaryotic ribosomes.
- Since mitochondria and chloroplasts have these ribosomes also, it is found in all 3 kingdoms.
- Most ribosomal RNA mutations are deleterious. Very few mutations are neutral.
- Therefore evolution of 16S RNA is very slow. It is a very good molecule to use to compare organisms that may have diverged as far back as 3 or 4 billion years ago.
- Visit the Ribosomal Database Project on the Web for access to data and software tools. This site lets users upload sequence information, generates alignments with other ribosomal genes, and returns aligned sequences with best matches, as well as generating phylogenetic trees.
But how do you actually do it?
- Originally would isolate large quantities of ribosomes, purify the small subunit, then purify the r-RNA, then sequence this.
- Most common method today is to use Polymerase Chain Reaction (PCR). PCR allows use of very small amounts of DNA as initial template. Using an appropriate primer to target desired region of DNA, can replicate this DNA again and again, with exponentially growing amounts of product.
- Three steps in PCR process:
- Denaturation: heat the DNA to around 94o C. This causes H-bonds holding A-T and G-C pairs to break, separation of two chains
- Primer Annealing: cool to 50o C. Aadd primer (short piece of RNA 10-20 nucleotides long) to denatured DNA. Incubate to allow formation of primer-DNA base pairing.
- Polymerization: warm to 72o C. Add DNA polymerase (isolated from thermophilic bacteria, so very heat-stable) plus dATP, dCTP, dGTP and dTTP.
- View animation of PCR cycling (requires Shockwave plug-in)
- Keep recycling these steps (all carried automatically inside closed box). Each time, double the amount of DNA: 1x --> 2x --> 4x --> 8x --> 16x --> 32x --> 64x .......... In 30 cycles, can get over 1 million molecules of product DNA!
- For 16S RNA studies, need to use primer that is found in every RNA molecule (universal primer), so probe will always find specific DNA site.
- Once sufficient DNA is produced, use DNA sequencing techniques to identify exact sequence.
The Bacterial Phylogenetic Tree
16S RNA results have produced remarkable results!
- The "tree of life" has been revolutionized by using small subunit RNA as the standard yardstick for evolutionary comparisons.
- Visit the Tree of Life Web site for a comprehensive, on-going global collaboration to create an accurate phylogeny for life on earth
- Three major evolutionary groups, each as different from each other
- Note: Archaea must have diverged from Bacteria as long ago as the origin of Eucaryotes
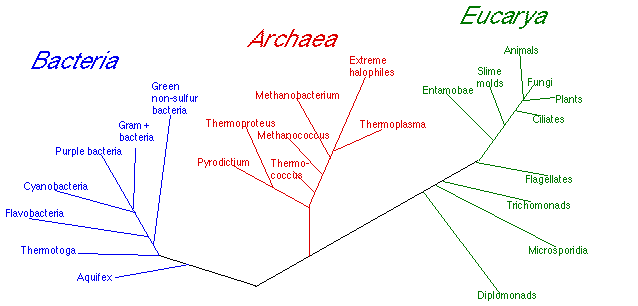
- This brings many differences between Bacteria and Archaea into perspective.
- Visit Major Groups of Prokaryotes page for further information (from Univ. of Wisconsin-Madison)
Bacteria consist of approximately 10 distinct groups
see MCB 409: Microbial Diversity at NC State University for more detail
- Gram + (including Mycoplasmas)
- Two major subdivisions:
- high G+C group (Actinomycetes, Mycobacteria, Micrococcus, others)
- low G+C group (Bacillus, Clostridia, Lactobacillus, Staphylococci, Streptococci, Mycoplasmas)
- For more information....
- Proteobacteria = Purple photosynthetic + non-photosynthetic Gram-negative
- includes most "common" Gram-negative bacteria
- For more information....
- ancestral species was a photosynthetic purple bacterium
- Cyanobacteria: oxygenic phototrophs
- Spirochetes and relatives
- Planctomyces
- Have never been cultivated, but common in pond water.
- Distant cousins of Chlamydia, also lack peptidoglycan
- Free-living aquatic oligotrophs; divide by budding, not binary fission.
- All have fimbriae & flagella.
- At least some have nuclear envelopes, like eukaryotes.
- Bacteroides, Flavobacteria
- intracellular parasites, no peptidoglycan
- Green sulfur bacteria
- anaerobic phototrophs
- Green non-sulfur bacteria
- also anaerobic phototrophs
- Radioresistant micrococci and relatives
- Very resistant to radiation, including gamma rays, X-rays, and UV. More than 20x as resistant as E. coli. Cells have very efficient DNA repair systems, multiple copies of DNA.
- Often show up in the spoilage of radiation-pasteurized food. Dose of gamma-rays used in food sterilization is very high precisely because of the need to kill Deinococcus. Compare with use of very high temperatures to preserve food, because of need to kill heat-resistants spores.
- Thermotoga and Thermosipho
- thermophiles from marine vents
Archaea consist of 3 distinct groups
- Archaea means "ancient" because use ancient energy mechanisms
- Many found in harsh, early earth-like environments.
- Introduction to the Archaea (requires password to The Biology Place)
Major Archaeal groups
- Thermal vents at bottom of ocean
- extreme salt conditions (Great Salt Lake, Dead Sea)
- high acid conditions.
- halophiles
- Example: Halobacterium --
- Found only in very concentrated brines, evaporating salt basins, Dead Sea, etc.
- Brightly colored due to purple pigments (bacteriorhodopsin)
- Use light energy to pump protons across cell membrane, generate proton gradient; make ATP from this
- methanogens
- Example: Methanococcus jannaschii , the first archaeal organism to have its genome sequenced
- extreme thermophiles
- Example: Pyrococcus furiosis; grows well at temperatures above boiling!
Web sites for further exploration: Microbial Diversity course at North Carolina State University
Take a Self-Quiz on this material
Return to Lecture Index
Return to MCB 229 Course Resources page